Data IO (input/output)¶
Introduction¶
ESRF data (used to) come in (too many) different formats:
- Specfile, EDF, HDF5
- And specific detector formats: MarCCD, Pilatus CBF, Dectris Eiger, …
HDF5 is now the standard ESRF data format so we will only focus on it today.
Methods for accessing other file formats are described in the io_spec_edf.ipynb notebook.
HDF5¶
What is HDF5?¶
HDF5 (for Hierarchical Data Format) is a file format to structure and store complex and large volumes of data.
Why HDF5?¶
- Hierarchical collection of data (directory and file, UNIX-like path)
- High-performance (binary)
- Portable file format (Standard exchange format for heterogeneous data)
- Self-describing extensible types, rich metadata
- Support data compression
- Free ( & open source)
- Adopted by a large number of institutes (NASA, LIGO, ...)
- Adopted by most of the synchrotrons (ESRF, SOLEIL, Desy...)
Data can be mostly anything: image, table, graphs, documents
HDF5 description¶
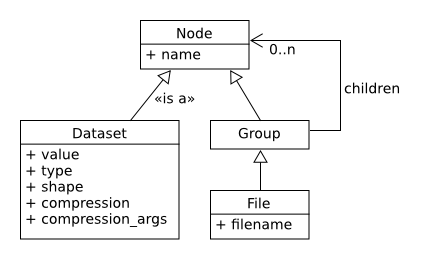
The container is mostly structured with:
- File: the root of the container
- Group: a grouping structure containing groups or datasets
- Dataset: a multidimensional array of data elements
- And other features (links, attributes, datatypes, virtual datasets)
Useful tools for HDF5¶
HDFGroup tools¶
Command line and desktop application: h5ls, h5dump, hdfview
>>> h5ls -r my_first_one.h5
/ Group
/data1 Dataset {100, 100}
/group1 Group
/group1/data2 Dataset {100, 100}
# From jupyter notebook
from h5glance import H5Glance
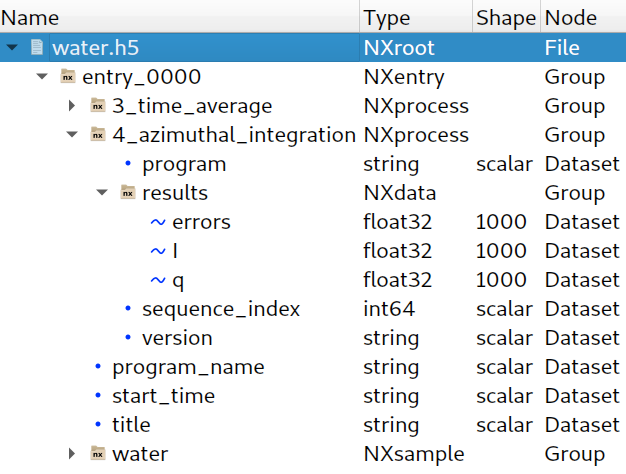
H5Glance("data/water.h5")
%%bash
# From the command line
h5glance data/water.h5
from jupyterlab_h5web import H5Web
H5Web("data/water.h5")
%%bash
# With silx view GUI
silx view data/water.h5
import h5py
print("h5py:", h5py.version.version)
print("hdf5:", h5py.version.hdf5_version)
How to read a HDF5 file from Python¶
h5py.File¶
First open the file with h5py.File:
h5py.File('myfile.hdf5', mode)
| Mode | Meaning |
|---|---|
| r | Readonly, file must exist; Default with h5py v3 |
| r+ | Read/write, file must exist |
| w | Create file, truncate if exists |
| w- or x | Create file, fail if exists |
| a | Read/write if exists, create otherwise; Default with h5py v2 |
import h5py
h5file = h5py.File("data/water.h5", mode="r")
h5py.Group¶
Then access the file content with a dictionary-like API, h5py.Group:
# Available names at the first level
list(h5file.keys())
# List 'entry_0000' group children
group = h5file["entry_0000"]
dict(group.items())
# List 'entry_0000/4_azimuthal_integration' group children
list(group["4_azimuthal_integration"].values())
# List 'entry_0000/4_azimuthal_integration/results' group children
list(h5file["/entry_0000/4_azimuthal_integration/results"].values())
# Get a dataset from a sub group
h5dataset = h5file["/entry_0000/4_azimuthal_integration/results/I"]
h5dataset
Not very convenient for interactive browsing... this is why silx view, h5web, h5glance ... exists
h5py.Dataset¶
It mimics numpy.ndarray.
The data is read from the file only when it is needed.
h5dataset = h5file["/entry_0000/4_azimuthal_integration/results/I"]
h5dataset
# Here we only read metadata from the dataset
print("Dataset:", h5dataset.shape, h5dataset.dtype, h5dataset.size)
# Read data from the file to a numpy.ndarray
subset = h5dataset[:5] # Copy the selection to a numpy.ndarray
print("subset:", subset, "=> sum:", subset.sum())
data = h5dataset[()] # Copy the whole dataset to a numpy.ndarray
print("data type:", type(data), "; shape", data.shape, "; min.:", data.min())
data[:5] = 0
print(data[:5])
print(h5dataset[:5])
# Remember to close the file
h5file.close()
print(data[:5])
print(subset)
# Once the file is closed, the Dataset no longer gives access to data
print(h5dataset)
print(h5dataset[:5])
Context manager¶
- Context managers guarantee that resources are released. In our case, it ensures that the HDF5 file is closed.
- Usually used from the
withstatement.
To safely access a HDF5 file, do:
with h5py.File("data/water.h5", "r") as h5file:
dataset = h5file["/entry_0000/4_azimuthal_integration/results/I"]
data = dataset[()]
print(dataset)
print(data[:5])
import numpy
import h5py
data = numpy.random.random(10000)
data.shape = 100, 100
# create file, notice the mode='w', as 'write'
h5file = h5py.File("my_first_one.h5", mode="w")
# write data into a dataset from the root
h5file["/data1"] = data
# write data into a dataset from group1
h5file["/group1/data2"] = data
h5file.close()
The same operation with a context manager:
import numpy
import h5py
# Create 2D data
data = numpy.arange(100 * 100)
data.shape = 100, 100
with h5py.File("my_first_one.h5", mode="w") as h5file:
# write data into a dataset from the root
h5file["/data1"] = data
# write data into a dataset from group1
h5file["/group1/data2"] = data
With Group.create_dataset and Group.create_group (alternative APIs) :
import numpy
import h5py
# Create 2D data
data = numpy.arange(100 * 100)
data.shape = 100, 100
# Notice the mode='w', as 'write'
with h5py.File("my_first_one.h5", mode="w") as h5file:
# write data into a dataset from the root
h5file.create_dataset("data1", data=data)
# Or with a functional API
grp1 = h5file.create_group("group1")
grp1.create_dataset("data2", data=data)
Exercice: Flat field correction¶
Flat-field correction is a technique used to improve quality in digital imaging.
The goal is to normalize images and remove artifacts caused by variations in the pixel-to-pixel sensitivity of the detector and/or by distortions in the optical path. (see https://en.wikipedia.org/wiki/Flat-field_correction)
$$ normalized = \frac{raw - dark}{flat - dark} $$
normalized: Image after flat field correctionraw: Raw image. It is acquired with the sample.flat: Flat field image. It is the response given out by the detector for a uniform input signal. This image is acquired without the sample.dark: Also namedbackgroundordark current. It is the response given out by the detector when there is no signal. This image is acquired without the beam.
Here is a function implementing the flat field correction:
Note: make sure you execute the cell for defining this function
import numpy
def flatfield_correction(raw, flat, dark):
"""
Apply a flat-field correction to a raw data using a flat and a dark.
"""
# Make sure that the computation is done using float
# to avoid type overflow or loss of precision
raw = raw.astype(numpy.float32)
flat = flat.astype(numpy.float32)
dark = dark.astype(numpy.float32)
# Do the computation
return (raw - dark) / (flat - dark)
Note: If you like to plot an image you can use matplotlib's imshow function.
The %matplotlib "magic" command should be called once first.
%matplotlib inline
from matplotlib import pyplot as plt
import numpy
plt.imshow(numpy.random.random((20, 60)))
Exercise 1¶
- Browse the file
data/ID16B_diatomee.h5 - Get a single raw dataset, a flat field dataset and a dark image dataset from this file
- Apply the flat field correction
- Save the result into a new HDF5 file
If you are stuck, the solution is provided in the file solutions/exercise1.py
from jupyterlab_h5web import H5Web
H5Web("data/ID16B_diatomee.h5")
# or
from h5glance import H5Glance
H5Glance("data/ID16B_diatomee.h5")
import h5py
with h5py.File("data/ID16B_diatomee.h5", mode="r") as h5s:
pass
# this is a comment
# step1: Read the data
# raw_data_path = ...
# raw_data = ...
# flat_path = ...
# flat = ...
# dark_path = ...
# dark = ...
# step2: Compute the result
# normalized = flatfield_correction(raw_data, flat, dark)
# step3: Save the result
# ...
Exercise 2¶
- Apply the flat field correction to all raw data available (use the same flat and dark for all the images)
- Save each result into different datasets of the same HDF5 file
If you are stuck, the solution is provided in the file solutions/exercise2.py
Exercise 3¶
From the previous exercise, we can see that the flat field correction was not very good for the last images.
Another flat field was acquired at the end of the acquisition.
We could use this information to compute a flat field closer to the image we want to normalize. It can be done with a linear interpolation of the flat images by using the name of the image as the interpolation factor (which varies between 0 and 500 in this case).
- For each raw data, compute the corresponding flat field using lineal interpolation (between
flatfield/0000andflatfield/0500) - Save each result into different datasets in a single HDF5 file
If you are stuck, the solution is provided in the file solutions/exercise3.py
Advanced usage¶
Dataset compression¶
Install hdf5plugin and import hdf5plugin.
HDF5 provides dataset compression support.
With h5py GZIP and LZF compression are available (see compression-filters).
Yet, there are many third-party compression filters for HDF5 available.
hdf5plugin allows usage of some of those compression filters with h5py (Blosc, Blosc2, BitShuffle, BZip2, FciDecomp, LZ4, SZ, SZ3, Zfp, ZStd).
import h5py
import hdf5plugin # Allows to read dataset stored with supported compressions
To write compressed datasets, see:
- Group.create_dataset
chunks,compressionandcompression_optsparameters. - "Chunked Storage" documentation
- hdf5plugin documentation
Soft and external links¶
A HDF5 file can contain links to Group/Dataset:
- within the same file: see h5py.SoftLink
- in another file: see h5py.ExternalLink
Links can be dangling if the destination does not exist.
External dataset¶
A HDF5 file can contain datasets that are stored in external binary files: See Group.create_dataset external parameter.
Virtual Dataset (aka. VDS)¶
Virtual dataset allows to map multiple datasets into a single one. Once created it behaves as other datasets.
HDF5 file locking: A word of caution¶
Do NOT open a HDF5 file that is otherwise being written (without caution).
By default, HDF5 locks the file even for reading, and other processes cannot open it for writing. This can be an issue, e.g., during acquisition.
WARNING: Without file locking, do not open twice the same file for writing or the file will be corrupted.
Workarounds:
- Helper to handle HDF5 file locking:
silx.io.h5py_utils.File - HDF5 file locking can be disabled by setting the
HDF5_USE_FILE_LOCKINGenvironment variable toFALSE. - With recent version of
h5py(>= v3.5.0):h5py.File'slockingargument
Practical tools¶
conversion:
silx convert: To convert EDF, or spec files to HDF5
reading/writing HDF5 helpers:
silx.io.dictdump:h5todict,dicttoh5silx.io.utils.h5py_read_dataset
A word about Nexus¶
Nexus is a data format for neutron, x-ray, and muon science.
It aims to be a common data format for scientists for greater collaboration.
If you intend to store some data to be shared it can give you a 'standard way' for storing it.
The main advantage is to ensure compatibility between your data files and existing softwares (if they respect the nexus format) or from your software to different datasets.
- an example on how to store tomography raw data
- an example to store tomoraphy application (3D reconstruction)
Conclusion¶
h5py provides access to HDF5 file content from Python through:
h5py.Fileopens a HDF5 file:- Do not forget the mode:
'r','a','w'. - Use a
withstatement or do not forget toclosethe file.
- Do not forget the mode:
h5py.Groupprovides a key-value mappingdict-like access to the HDF5 structure.h5py.Datasetgives access to data asnumpy.ndarray.